Introduction
Generalized median strings have been successfully applied to many pattern recognition problems. In many situations, noise often ends up the culprit of recognition failure. Median strings can help by averaging the results and finding the best estimate among many noisy samples. It is also possible to use median string algorithms to compute among a set of noisy samples the best prototypes for machine learning.
The nice properties of generalized median strings are useful in diverse areas for many applications such as symmetry analysis, shape recognition, optical character recognition, bar code reading, speech recognition, molecular biology, other pattern recognitions problems, signal estimation in multisensory measurements, clustering, and averaging of many kinds of data, among others: binary feature maps, 3D rotations, other geometric features, brain models, anatomical structures, and facial images.
In this tutorial, we will first define what is a string and how to compute the distance or difference between two strings. Next, we will extend the concept by defining weighted means of strings which will allow us among other things to compute strings that are halfway between a pair of strings. Then, we will introduce the concept of generalized median strings, the goal of this tutorial. Finally, we will describe a concrete application to 2D shapes, and let you play with a fun applet that will let you draw your own shapes and have the computer compute median shapes.
I, Samuel Audet, made this tutorial as my final course project for COMP 644 – Pattern Recognition at McGill University in the fall term of 2006, a course given by Professor Godfried Toussaint.
Strings
So, what is a string? A string is a list of symbols. Symbols can be, for example, letters, numbers or
vectors, but any arbitrary finite alphabet of
symbols can be used. 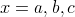 and
and
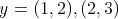 are two examples of
strings. The first uses letters as symbols and has three of them, so its length equals 3. The second
uses two-dimensional vectors and since it contains
two vectors, its length equals 2.
are two examples of
strings. The first uses letters as symbols and has three of them, so its length equals 3. The second
uses two-dimensional vectors and since it contains
two vectors, its length equals 2.
Edit operations
Now, we can perform what we will call edit operations on a string. There exists three kinds of operations and we call them insertions, substitutions, and deletions, as illustrated in figure 1.
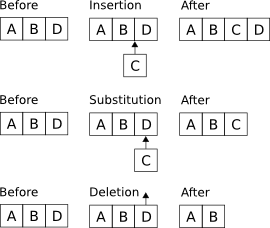
Figure 1. Illustration of the three possible edit operations: insertions, substitutions and deletions.
The operations in figure 1 are denoted as follows:
| Edit operation |  = = |
|---|---|
| Insertion of C | 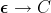 |
| Substitution of D with C |  |
| Deletion of D |  |
Note that the length of a string is increased by one after an insertion, reduced by one after a deletion, and remains unchanged after a substitution.
By definition, an edit operation has a cost equal to or greater than 0. The cost is denoted
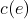 where
where  is an edit operation. When no changes occurs as the result of an edit operation, it is
called an identical substitution. In such cases, the cost of the operation must
be 0. Also, the cost function must follow the triangle
inequality:
is an edit operation. When no changes occurs as the result of an edit operation, it is
called an identical substitution. In such cases, the cost of the operation must
be 0. Also, the cost function must follow the triangle
inequality:
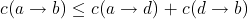
That is, in cases where one edit operation gives the same result as two other edit operations, the cost of the former
must always be smaller or equal to the sum of the costs of the latter. We can of course have a sequence of edit operations
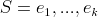 , and the cost of this sequence is simply the sum of the
costs of the edits, in other words:
, and the cost of this sequence is simply the sum of the
costs of the edits, in other words:
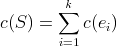
Edit distance
Now, we are ready to define the edit distance between two strings. The edit distance between two strings is important because
it tells us by how much two strings differ from one another. Also, the algorithm used will allow us to find an optimal edit sequence
that can transform a string into another. Before that though, these two strings that we will denote as 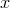 and
and 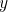 must belong to the same space, in other words, they must use the same alphabet of symbols.
One popular definition of their distance is equal to the minimum cost over all possible sequences
must belong to the same space, in other words, they must use the same alphabet of symbols.
One popular definition of their distance is equal to the minimum cost over all possible sequences
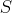 of edit operations that can change
of edit operations that can change 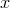 into
into 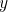 , in other words:
, in other words:
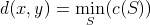
This is commonly known as the Levenshtein edit distance.
The assumption of the first inequality also implies the triangle inequality for string distances:
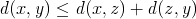
There exists a standard algorithm using dynamic programming
to compute the distance between two strings
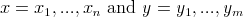 .
This algorithm is described by the recursive relationships of the following equations:
.
This algorithm is described by the recursive relationships of the following equations:
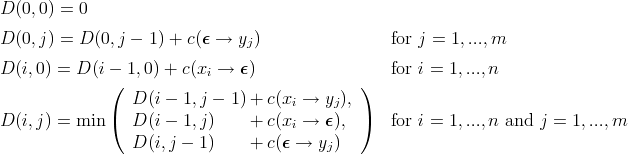
Wagner and Fischer [1] proved that 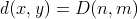 .
We can also extract from the path taken in the matrix an
optimal edit sequence
.
We can also extract from the path taken in the matrix an
optimal edit sequence  which has minimal cost.
which has minimal cost.
Here is a small example of the algorithm in action on two smalls strings
 and
and 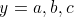 so that n = 2 and m
= 3. For simplicity, all edit operations have a cost of 1, except identical substitutions which must have a cost of 0.
The dynamic programming algorithm described above will compute the
matrix D as shown in figure 2. We can see that
so that n = 2 and m
= 3. For simplicity, all edit operations have a cost of 1, except identical substitutions which must have a cost of 0.
The dynamic programming algorithm described above will compute the
matrix D as shown in figure 2. We can see that 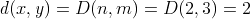 .
This is in fact true since to transform
.
This is in fact true since to transform 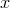 into
into
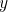 , we can substitute the 'd' with 'b' and insert a 'c', or we can insert a 'b' and substitute the 'd' with 'c'. Actually, the
former sequence of operations is depicted with the red line, and the latter, the blue line. This can easily be understood by
realizing that diagonal paths into the matrix are substitution operations. So, in the case of the red line, we have a
substitution of 'a' with 'a' (cost 0), then a substitution of 'd' with 'b' (cost 1), and finally, an insertion of 'c' (again,
cost 1, so total 2). Follow the same logic for the blue line as exercise.
, we can substitute the 'd' with 'b' and insert a 'c', or we can insert a 'b' and substitute the 'd' with 'c'. Actually, the
former sequence of operations is depicted with the red line, and the latter, the blue line. This can easily be understood by
realizing that diagonal paths into the matrix are substitution operations. So, in the case of the red line, we have a
substitution of 'a' with 'a' (cost 0), then a substitution of 'd' with 'b' (cost 1), and finally, an insertion of 'c' (again,
cost 1, so total 2). Follow the same logic for the blue line as exercise.
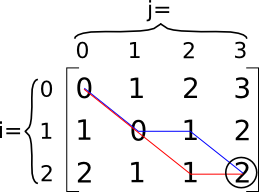
Figure 2. Illustration of the distance algorithm in action on two strings. This is the matrix D.
The blue and red lines are both optimal edit sequences that can transform string 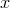 into string
into string 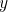 with minimal cost. It should be clear with this example that not even
an optimal edit sequence
with minimal cost. It should be clear with this example that not even
an optimal edit sequence 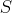 is necessarily unique.
is necessarily unique.
Weighted means of strings
In this section, we will talk about the weighted means of a pair of strings. This concept will guide us one step
further into our adventure in understanding generalized median strings. A weighted mean of a pair of strings
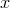 and
and 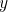 is a string
is a string
 that satisfies:
that satisfies:
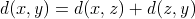
In other words, the distance between 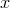 and
and 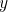 must be equal to the distance between
must be equal to the distance between 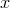 and the weighted mean
and the weighted mean  plus
the distance between, again, the weighted mean
plus
the distance between, again, the weighted mean  and
and 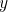 .
It is clear that
.
It is clear that  can be equal to either
can be equal to either 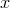 or
or 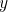 , so that
, so that 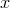 and
and 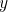 are
themselves weighted means as well. We can arbitrarily call
are
themselves weighted means as well. We can arbitrarily call 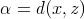 our weight. With this
definition, when
our weight. With this
definition, when 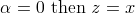 and when
and when 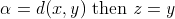 .
For intermediate values of
.
For intermediate values of 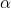 ,
,  is not necessarily unique.
For example, let us define two strings
is not necessarily unique.
For example, let us define two strings  and
and 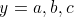 where
edit operations all have a cost of 1 for simplicity. With
where
edit operations all have a cost of 1 for simplicity. With 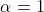 , we can think of two edit
operations that would bring us closer to
, we can think of two edit
operations that would bring us closer to 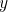 . They are insertions
. They are insertions
 and
and  .
These give us two different weighted means
.
These give us two different weighted means 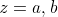 and
and
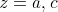 where
where 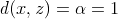 for both.
for both.
As we will see in the next section, a weighted mean is also usually a generalized median of a set of two strings.
To find a weighted mean at distance 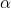 from
from
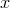 , we can use the following procedure:
, we can use the following procedure:
- Compute the distance
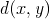 to find an optimal edit sequence
to find an optimal edit sequence 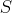 .
.
- Take a subset
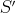 of operations from
of operations from 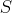 whose cost
whose cost 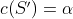 , assuming such a subset exists.
, assuming such a subset exists.
- Apply the operations of
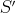 to
to 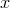 to produce
to produce  , a weighted mean.
, a weighted mean.
Here is a proof that this algorithm is correct. Since 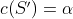 , then
, then
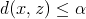 , since there might be a sequence
other than
, since there might be a sequence
other than 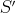 that is cheaper and that can transform
that is cheaper and that can transform 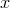 to
to
 . Then, since we do not need to apply the operations in
. Then, since we do not need to apply the operations in 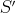 to
transform
to
transform  into
into 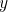 , the distance between
, the distance between  and
and 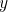 is reduced by at least that much, i.e.:
is reduced by at least that much, i.e.: 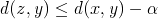 . From these two inequalities, we now know that
. From these two inequalities, we now know that 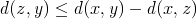 or alternatively
or alternatively 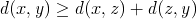 .
But we also know that
.
But we also know that 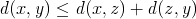 , the triangle inequality.
So it must be that
, the triangle inequality.
So it must be that 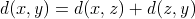 .
. 
Now, a subset 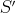 with cost
with cost 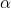 might not
exist, but in that case Bunke and others [2] proved that if we can decompose edit operations into
sub-operations, we still arrive to the same conclusion.
might not
exist, but in that case Bunke and others [2] proved that if we can decompose edit operations into
sub-operations, we still arrive to the same conclusion.
Generalized median strings
We can finally start talking about generalized median strings! Just before that however, we have to introduce the
notion of Sum Of Distances (SOD). Given a set of strings 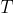 and an additional string
and an additional string
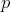 the Sum Of Distances (SOD) is the sum of distances from string
the Sum Of Distances (SOD) is the sum of distances from string 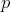 to all strings
to all strings 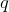 in set
in set 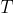 , in other words:
, in other words:
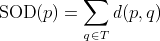
Assuming the strings are part of an arbitrary space 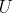 , a string
, a string
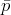 that belongs to
that belongs to 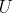 and that has
the smallest Sum Of Distances (SOD) is called a generalized median string, or in other words:
and that has
the smallest Sum Of Distances (SOD) is called a generalized median string, or in other words:
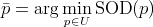
Similarly, a string 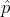 in the set
in the set 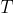 with the smallest Sum Of Distances (SOD) is called a set median, or in other words:
with the smallest Sum Of Distances (SOD) is called a set median, or in other words:
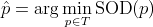
As we have seen, the weighted means of strings are not necessarily unique, so neither are median strings. Moreover, the
concept of generalized median strings is equivalent to the concept of weighted means of a pair of strings under some conditions.
If we assume that the cost function for a given problem is symmetric, i.e.: 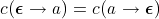 and
and 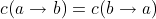 for any given
for any given
 and
and 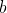 , then the distance is also symmetric,
i.e.:
, then the distance is also symmetric,
i.e.: 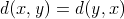 for any given
for any given 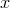 or
or
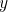 . In this case, it is clear that if the set
. In this case, it is clear that if the set 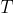 contains only two strings, the notion of generalized medians reduces to the notion of weighted means, since the
contains only two strings, the notion of generalized medians reduces to the notion of weighted means, since the
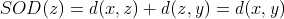 . In addition, note that the SOD does not actually
depend on
. In addition, note that the SOD does not actually
depend on  so it is equal for any weighted mean string
so it is equal for any weighted mean string
 , including
, including 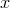 and
and
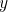 themselves.
themselves.
In general, however, we are interested in much larger sets of strings. The generalized median is the more general concept and therefore provides a better representation of the set than the set median, but de la Higuera and Casacuberta [3] proved that the computation of the former is NP-complete, in other words very hard to compute. On the other hand, the computation of the set median string is very straightforward. We can apply an algorithm directly modeled after the equation above, which suggests the following: Compute the Sum Of Distances (SOD) for all the strings in the set, and a string that gives the smallest SOD is a set median. This brute-force approach has time complexity O(n2) in the number of strings in the set. Computing the generalized median string is much harder and more often than not, only approximations can be computed. Case in point, in the applet below, the heuristic we used to compute an approximation is the dynamic computing approach by Jiang and others [4]. You will notice that under some circumstances it produces worse estimates (higher SOD) than the set median algorithm! The authors have also tried a genetic algorithm approach [5], which is also quite experimental. Casacuberta and de Antoni [6] have for their part taken a greedy approach. Those heuristics are all quite complex and mostly empirical, so they provide little insight into the issue of computing generalized median strings. In any case, we invite the interested reader to consult those papers.
Application to shapes
We will now describe an application to (two-dimensional) shapes where we are interested in finding the median shapes. We will use a vectorial
representation where all vectors have the same length  (a constant, that could be for
example 1). We have depicted in figure 3 an S-like shape using such a representation. All vectors have the same length
(a constant, that could be for
example 1). We have depicted in figure 3 an S-like shape using such a representation. All vectors have the same length
 (You can imagine
(You can imagine  = 1). In this
representation, the tail of the next vector (e.g.:
= 1). In this
representation, the tail of the next vector (e.g.: 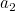 ) follows the head of the previous vector
(e.g.:
) follows the head of the previous vector
(e.g.: 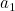 ), and so on.
), and so on.
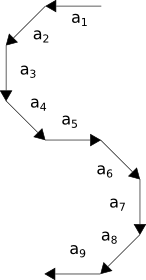
Figure 3. Illustration in vectorial form of a shape similar to an S where all nine vectors have the same length.
We can denote this shape in this representation as a string of vectors:
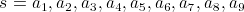 .
.
For this application, we define the costs of the edit operations as follows:
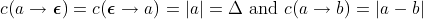 . In other words,
the cost of insertions and deletions are equal to the length of the vectors. Moreover,
. In other words,
the cost of insertions and deletions are equal to the length of the vectors. Moreover,  is
implied to be a Minkowski distance between two vectors. For example, we could use the
well known Euclidean distance:
is
implied to be a Minkowski distance between two vectors. For example, we could use the
well known Euclidean distance: 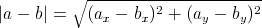 . With this definition
. With this definition 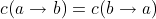 and
thus we end up with a symmetric distance metric for the whole string as discussed before.
Notice that the minimum cost is 0, and the maximum cost is
and
thus we end up with a symmetric distance metric for the whole string as discussed before.
Notice that the minimum cost is 0, and the maximum cost is 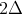 when
when  and
and
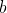 have the same orientation, but opposite directions, such as vectors
have the same orientation, but opposite directions, such as vectors
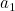 and
and 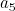 in the figure 3.
in the figure 3.
We can then apply the algorithms discussed previously to shapes represented with such strings. In particular, we can find a generalized median string of a set of shapes, which would give us in some perceptual way a "middle shape" of the set. For example, if we have many shapes of the letter S, they might look very similar to a human, but a machine does not know they are all supposed to be S's. We could tell the computer to find a generalized median string to help it find a prototype S. Since this single prototype has the minimal distance to shapes that we humans see as S's, it would make it much easier for the computer to recognize S's in the future, since it would only need to carry around the one prototype instead of the whole set. You will be able to play with the applet below and see what kind of prototypes the computer computes given 10 shapes that you can draw.
Fun applet
With this applet, you are allowed to draw up to 10 shapes and find out what the computer will decide are the median strings. Note that the shapes are limited to a single curve, so that you cannot draw shapes with disconnected components.
Next, the algorithm used to find an approximation of a generalized median string is the dynamic algorithm developed by Jiang and others [4]. It is dynamic in the sense that it computes an approximation of a generalized median string incrementally, one string at a time. Therefore, we get intermediate results of approximations computed for subsets of the whole set of strings. The applet exploits this fact by displaying the approximation as computed after each string is added to the computation. Watch for the blue background. Note that since this computation is only an approximation, we used randomness in the implementation, and this is why for the same set of shapes, the approximation is different each time the computation is launched.
The set median string is computed using the brute-force approach as described above. Watch for the green background.
Some interesting things to try with the applet:
- Draw 6 times the same character, and 4 other scribbles. Amazingly, the medians reflect the character drawn 6 times! But this is to be expected for something we call the median. Nonetheless, this demonstrates that the algorithms are robust to outliers.
- Try to draw this character 6 times again, but in different positions, scales and angles. If you are lucky, the character is still "discovered" by the median algorithms! The vectorial string representation is (more or less) insensitive to translation, scale and rotation.
- Use your imagination and try various things to see if they produce what you expect.
Note: If you do not see the applet that is supposed to appear here, make sure your browser is equipped with a Java Runtime Environment (JRE) of version 1.4.2 or later. Such JRE for Windows and Linux can be freely downloaded from Sun.
Download the source code of this applet.
References
| [1] | Robert A. Wagner and Michael J. Fischer, "The string-to-string correction problem," Journal of the ACM, vol. 21, no. 1, pp. 168–173, 1974. |
| [2] | Horst Bunke, Xiaoyi Jiang, Karin Abegglen and Abraham Kandel, "On the Weighted Mean of a Pair of Strings," Pattern Analysis and Applications, vol. 5, no. 1, pp. 23–30, 2002. |
| [3] | Colin de la Higuera and Francisco Casacuberta, "Topology of Strings: Median String is NP-Complete," Theoretical Computer Science, vol. 230, no. 1–2, pp. 39–48, 2000. |
| [4] | Xiaoyi Jiang, Karin Abegglen, Horst Bunke, and János Csirik, "Dynamic computation of generalised median strings," Pattern Analysis and Applications, vol. 6, no. 3, pp. 185–193, 2003. |
| [5] | Xiaoyi Jiang, Horst Bunke, and János Csirik, "Median Strings: A Review," Data Mining in Time Series Databases, World Scientific, pp. 173–192, 2004. |
| [6] | F. Casacuberta and M.D. de Antoni, "A greedy algorithm for computing approximate median strings," Proceedings of National Symposium on Pattern Recognition and Image Analysis, pp. 193–198, Barcelona, Spain, 1997. |