Firstly, This study was conducted in accordance with the Declaration of Helsinki. The Institutional Review
Board at Nihon University Hospital approved the study protocol on March 8, 2018, before patient recruitment.
Informed consent was obtained from all patients before they were enrolled. This study was registered with
the University Hospital Medical Information Network (UMIN) Clinical Trials Registry (identification No.:
UMIN000031776) on March 17, 2018. This study was also approved by the research ethics committee of Tokyo
Institute of Technology, where 3D reconstruction experiments were conducted.
In our previous study, we tackled the problem of lesion localization by reconstructing a whole stomach 3D
shape from endoscopic images based on structrure-from-motion (SfM) pipeline. Unfortunately, our previous
pipeline only works with gastroendoscopy sequence with indigo carmin blue dye. However, though the IC dye is
commonly used in gastroendoscopy, spraying it on the whole stomach surface requires additional procedure
time, labor, and cost, which is not desirable for both patients and practitioners. Furthermore, the IC dye
may also hinder the visibility for some lesions and reconstructed stomach 3D models because of its dark
color tone. In this work, we expand our previous work by proposing a framework to reconstruct the whole
stomach shape from endoscope video using SfM pipeline without the need of indigo carmin blue dye.
Based on the investigation, we have found that the generated VIC images significantly increase the number of
extracted SIFT feature points. We have also found that translating from no-IC green-channel images to
IC-sprayed red-channel images gives significant improvements to the SfM reconstruction quality. We have
experimentally demonstrated that our new approach can reconstruct the whole stomach shapes of all seven
subjects and showed that the estimated camera poses can be used for the lesion localization purpose.
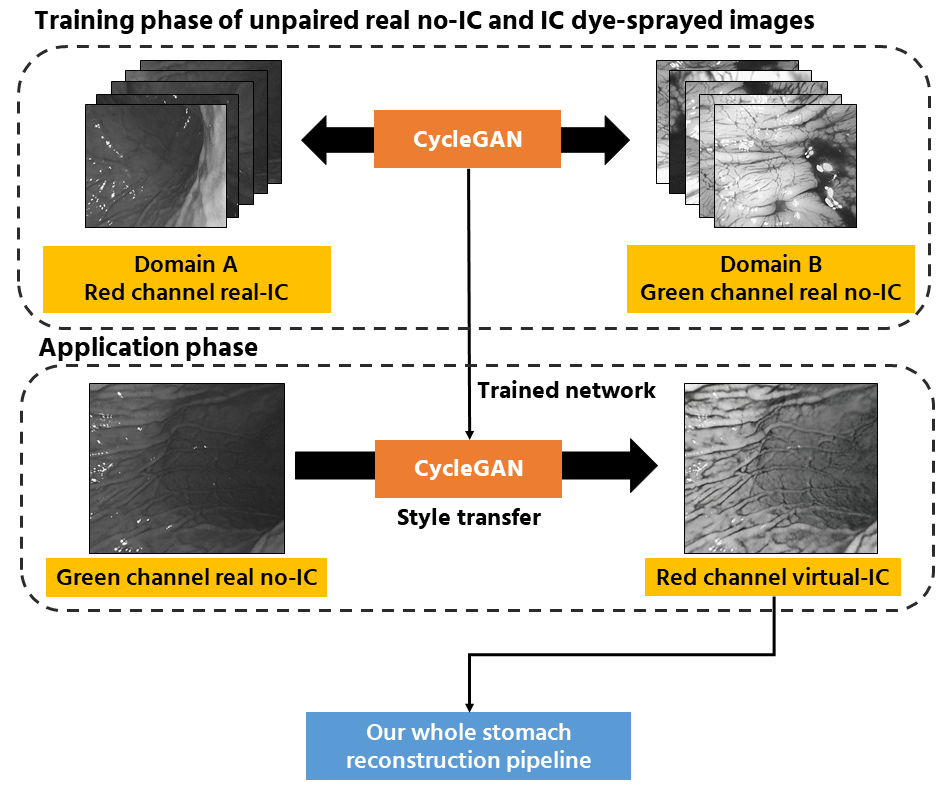 The flow of overall pipeline.
The flow of overall pipeline.
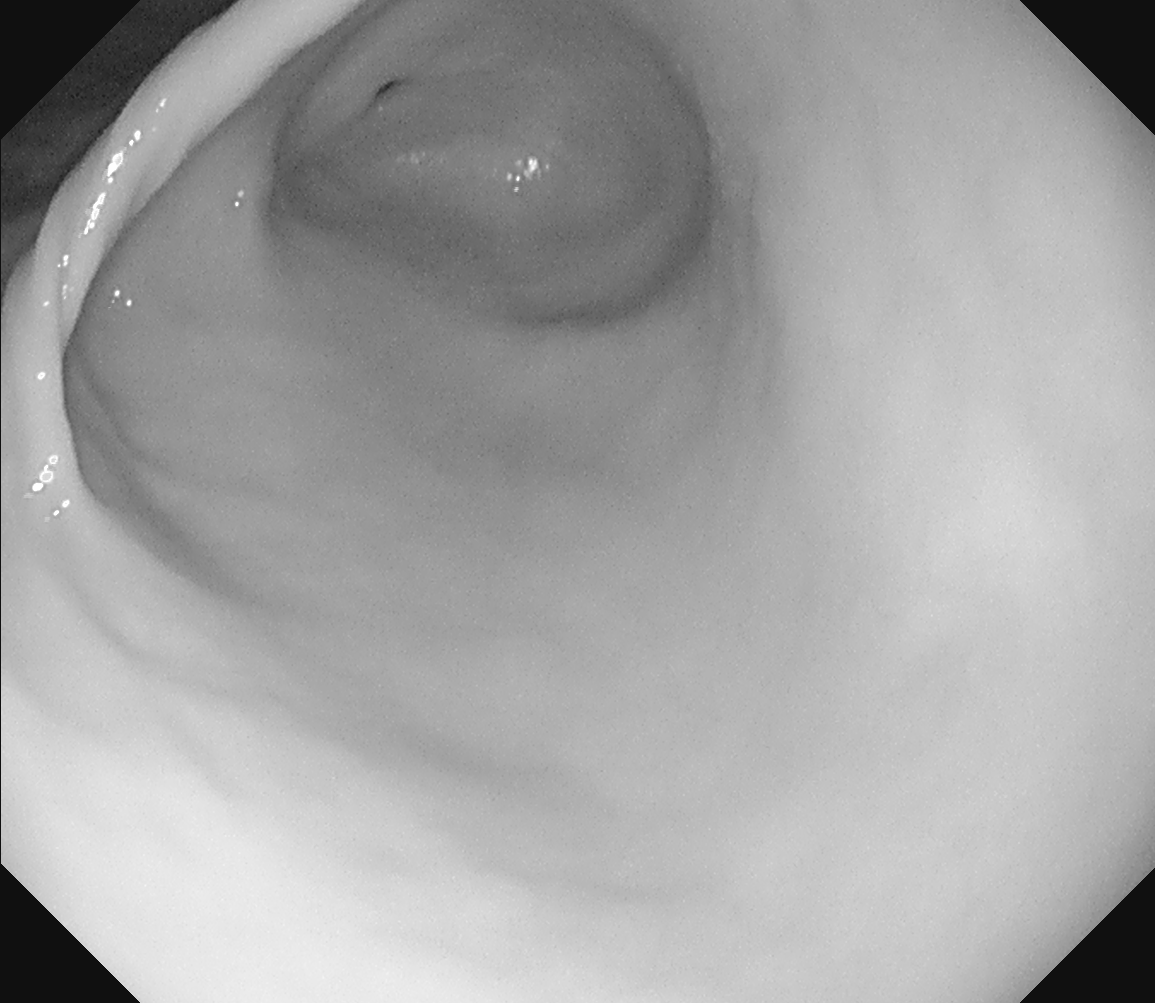 Real red channel no-IC
Real red channel no-IC
.png) Real red channel no-IC to virtual red channel with IC
Real red channel no-IC to virtual red channel with IC
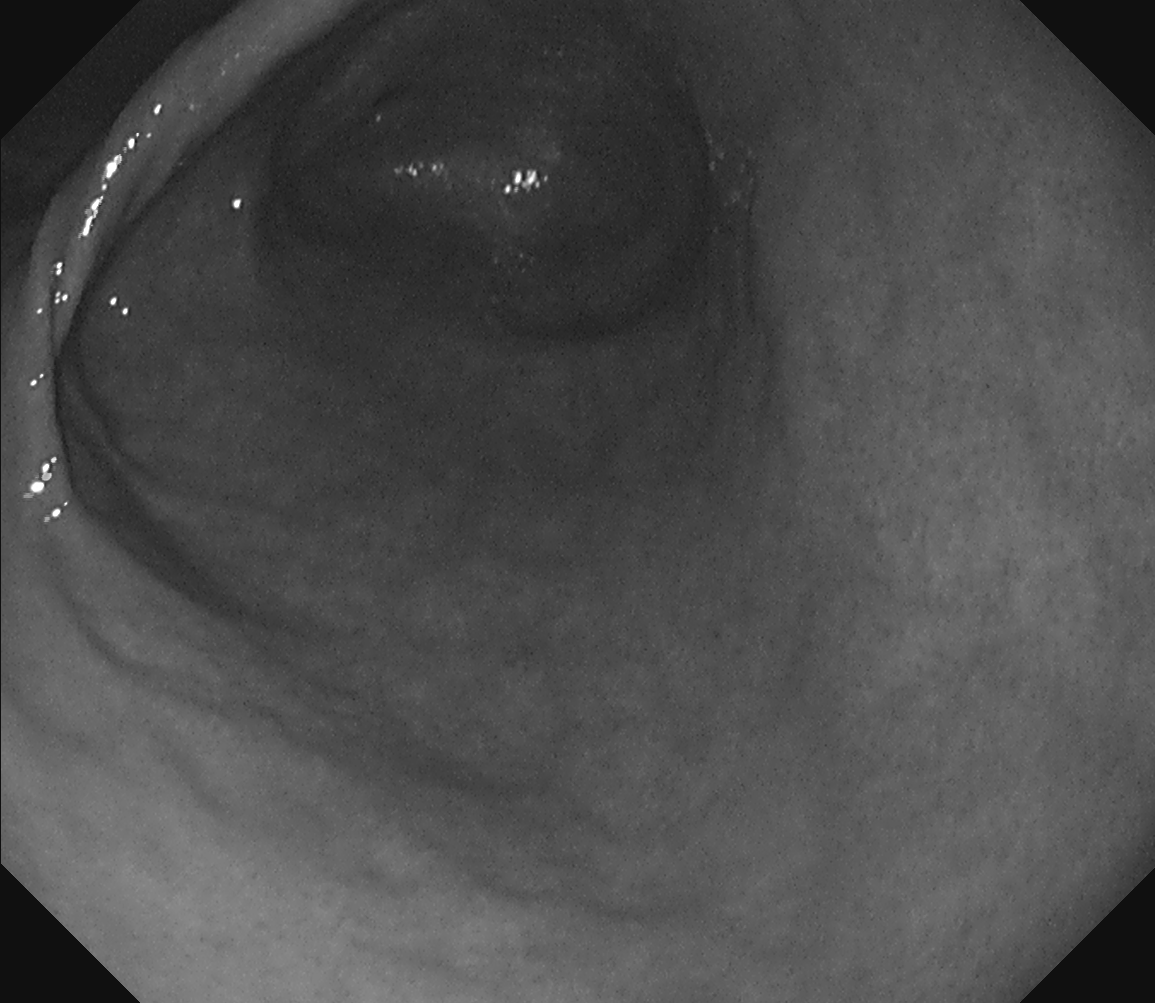 Real green channel no-IC
Real green channel no-IC
.png) Real green channel no-IC to virtual red channel with IC
Real green channel no-IC to virtual red channel with IC
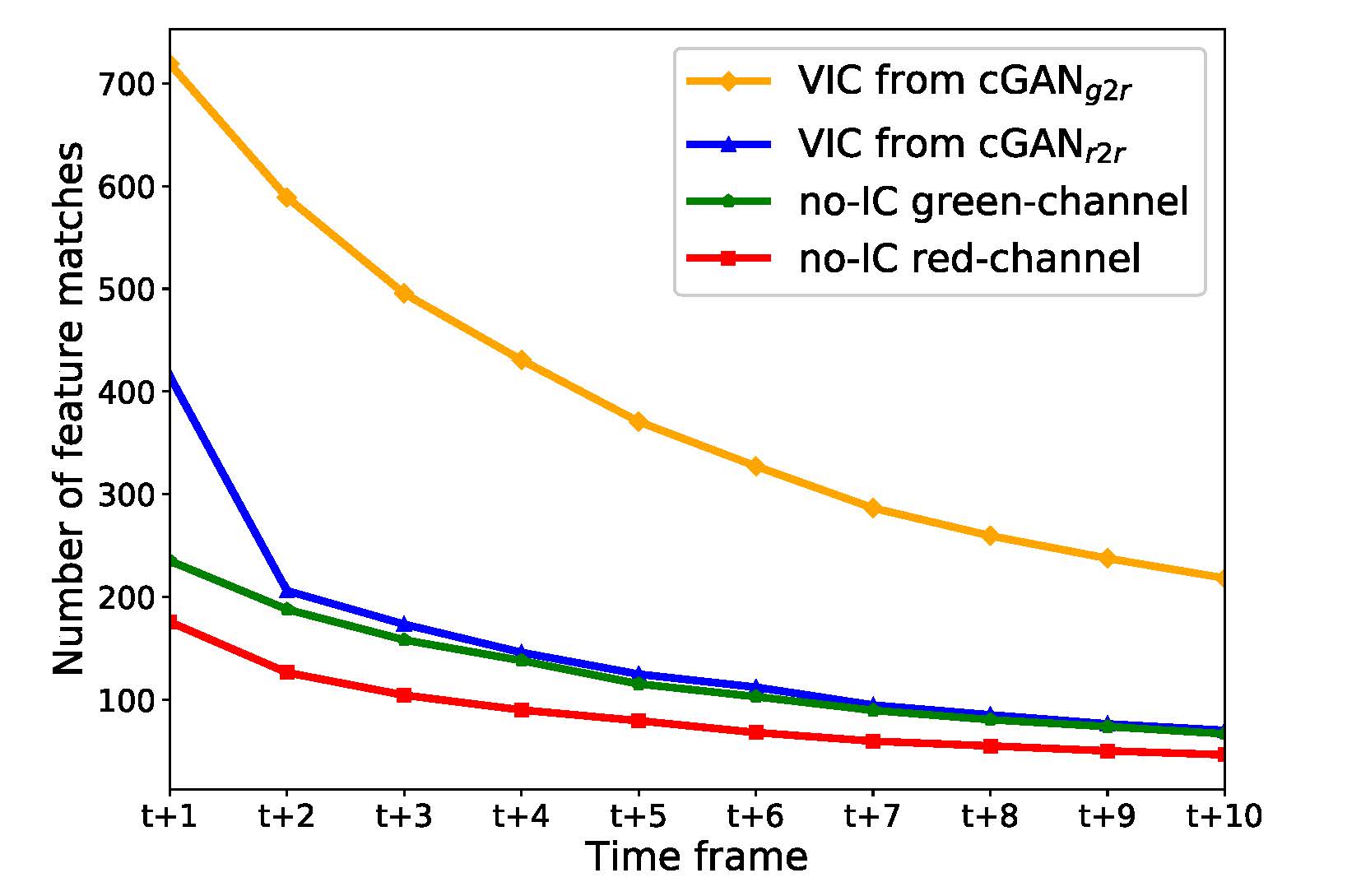
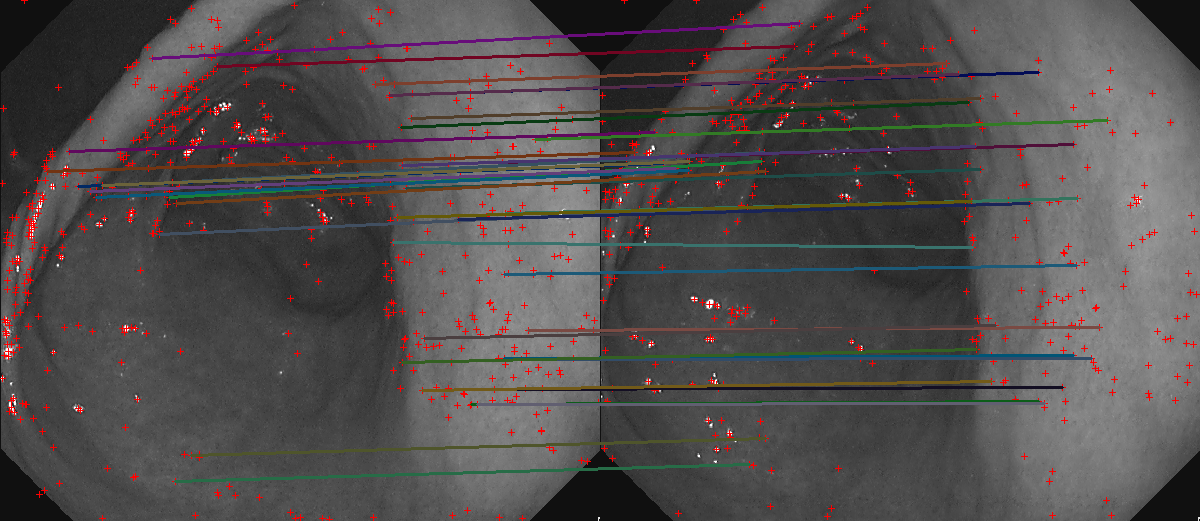 Feature matches on green channel no-IC images
Feature matches on green channel no-IC images
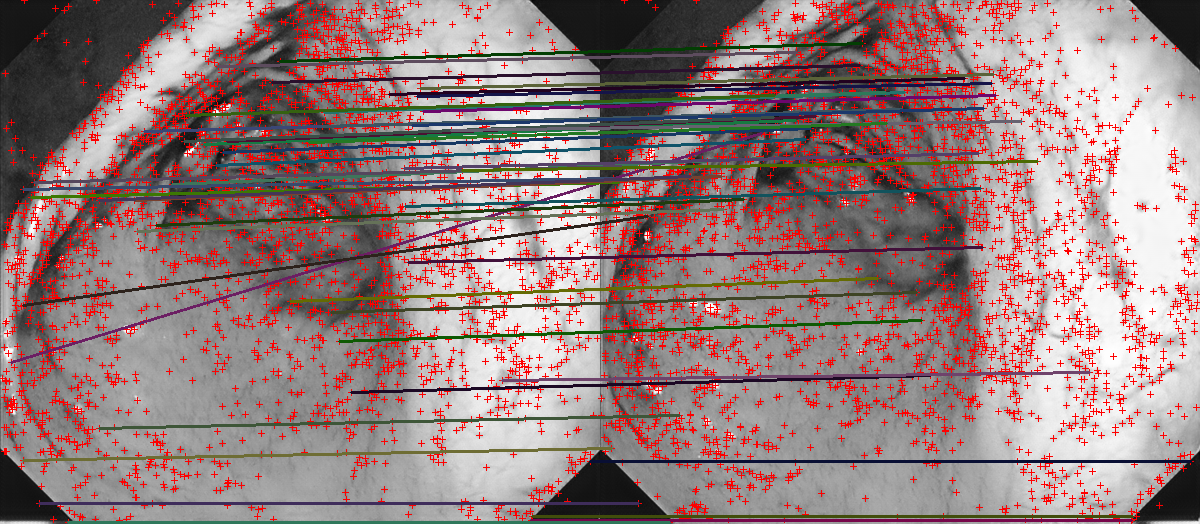 Feature matches on VIC images from cGANr2r
Feature matches on VIC images from cGANr2r
 Feature matches on VIC images from cGANg2r
Feature matches on VIC images from cGANg2r