Flowchart
This section illustrates the overall flow of our proposed pipeline.
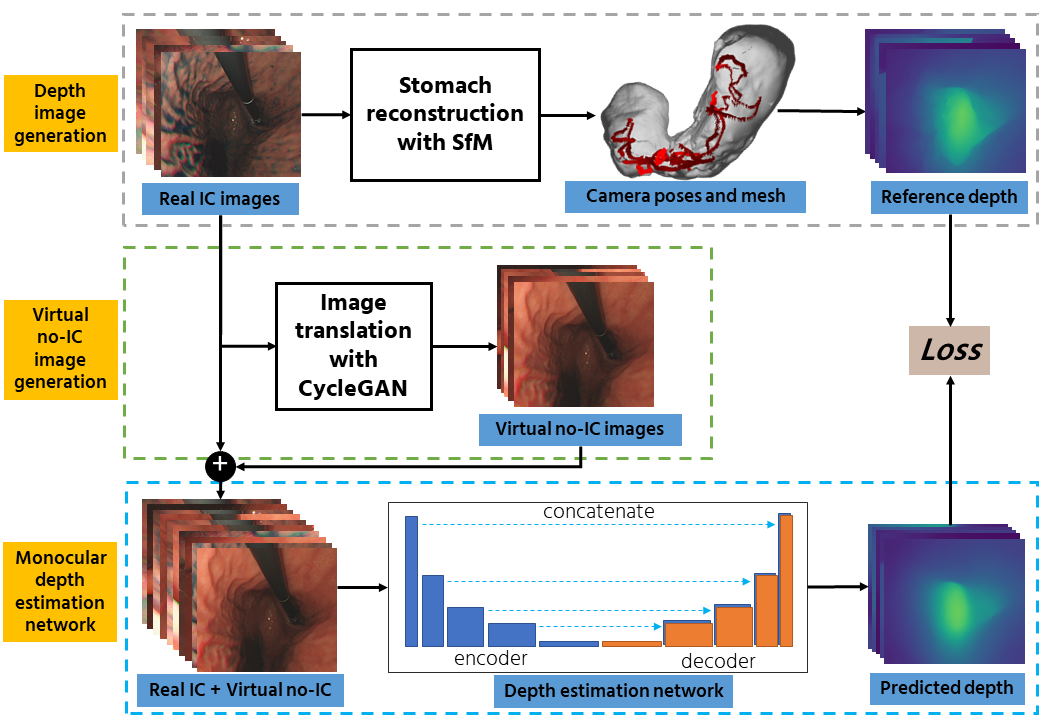 The overall flow of the proposed pipeline.
The overall flow of the proposed pipeline.
Our proposed method is divided into three main parts: (i). Reference depth data generation, (ii). Virtual no-IC image generation, and (iii). Monocular depth estimation training.
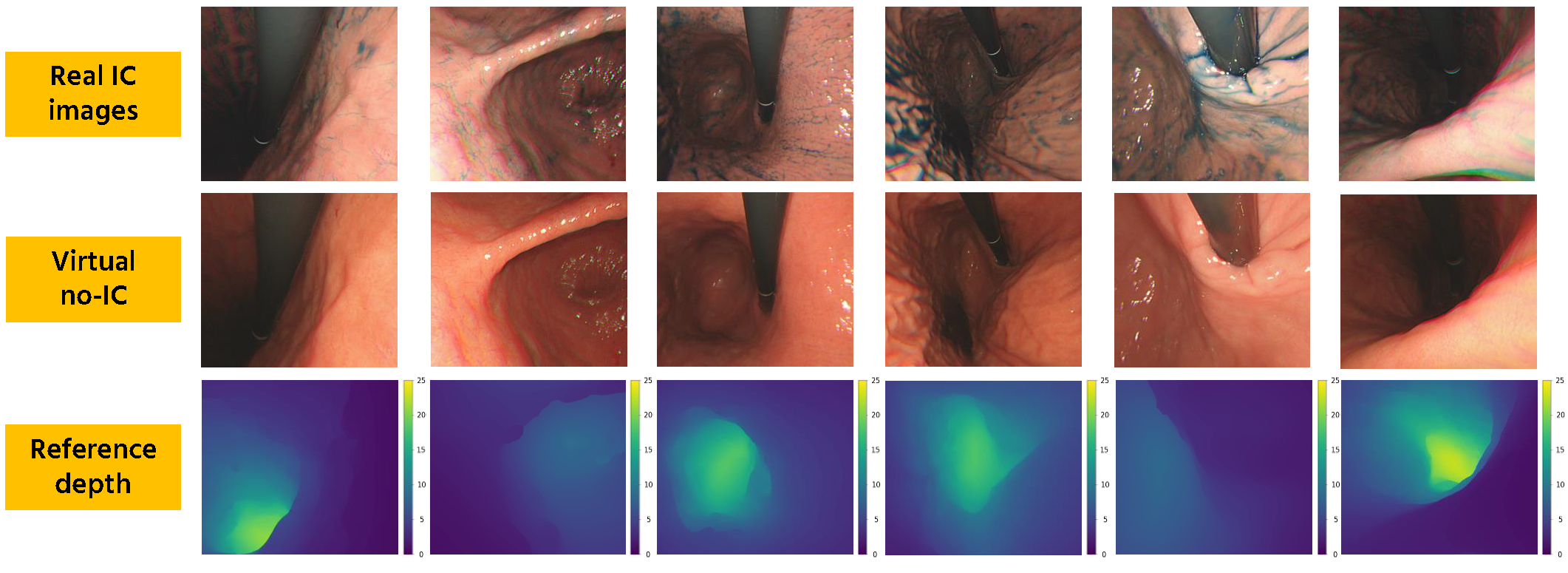
Reference depth data generation. We first apply a stomach 3D reconstruction SfM pipeline2 to estimate camera poses and reconstruct a 3D mesh model. Here, we use real IC-dye-sprayed texture-enhanced images (IC images) as SfM inputs since dense 3D reconstruction cannot be achieved using standard no-IC images due to its textureless surface properties. Using the obtained 3D mesh, we then generate dense reference depth for each reconstructed camera.
Virtual no-IC image generation. The dense stomach 3D reconstruction pipeline2 cannot work with no-IC images because of its textureless surface. Thus, reference depth data for no-IC images cannot be directly obtained, which limits the depth estimation to only real IC images. To address this issue, we propose to apply CycleGAN that works with unpaired data to generate virtual no-IC images. To train the CycleGAN, we used unpaired real IC and real no-IC images extracted from our experimental endoscope data. We then use the trained CycleGAN to generate virtual no-IC images. This approach enables us to create the pairs of reference depth images and no-IC images for self-supervised depth training.
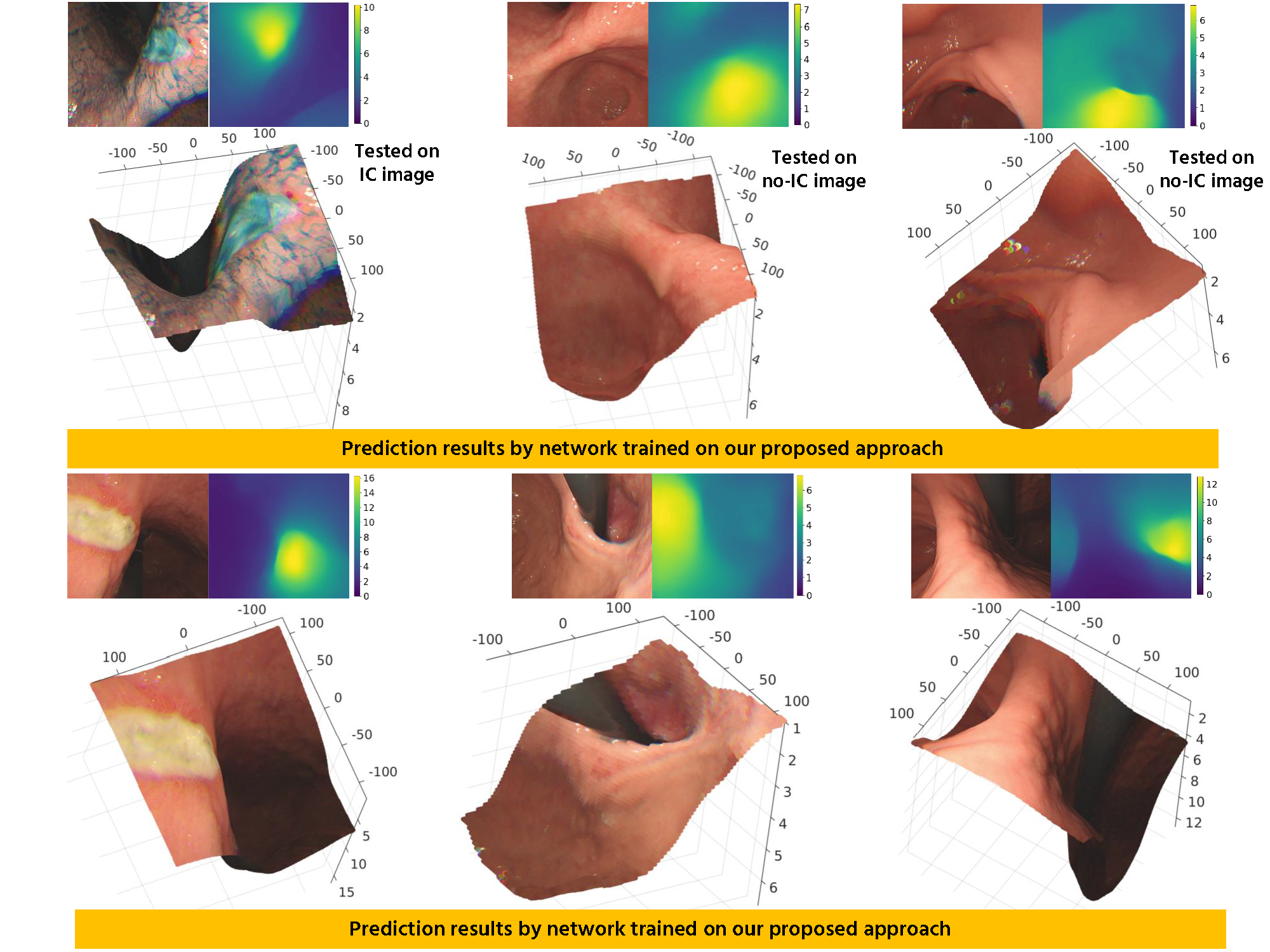
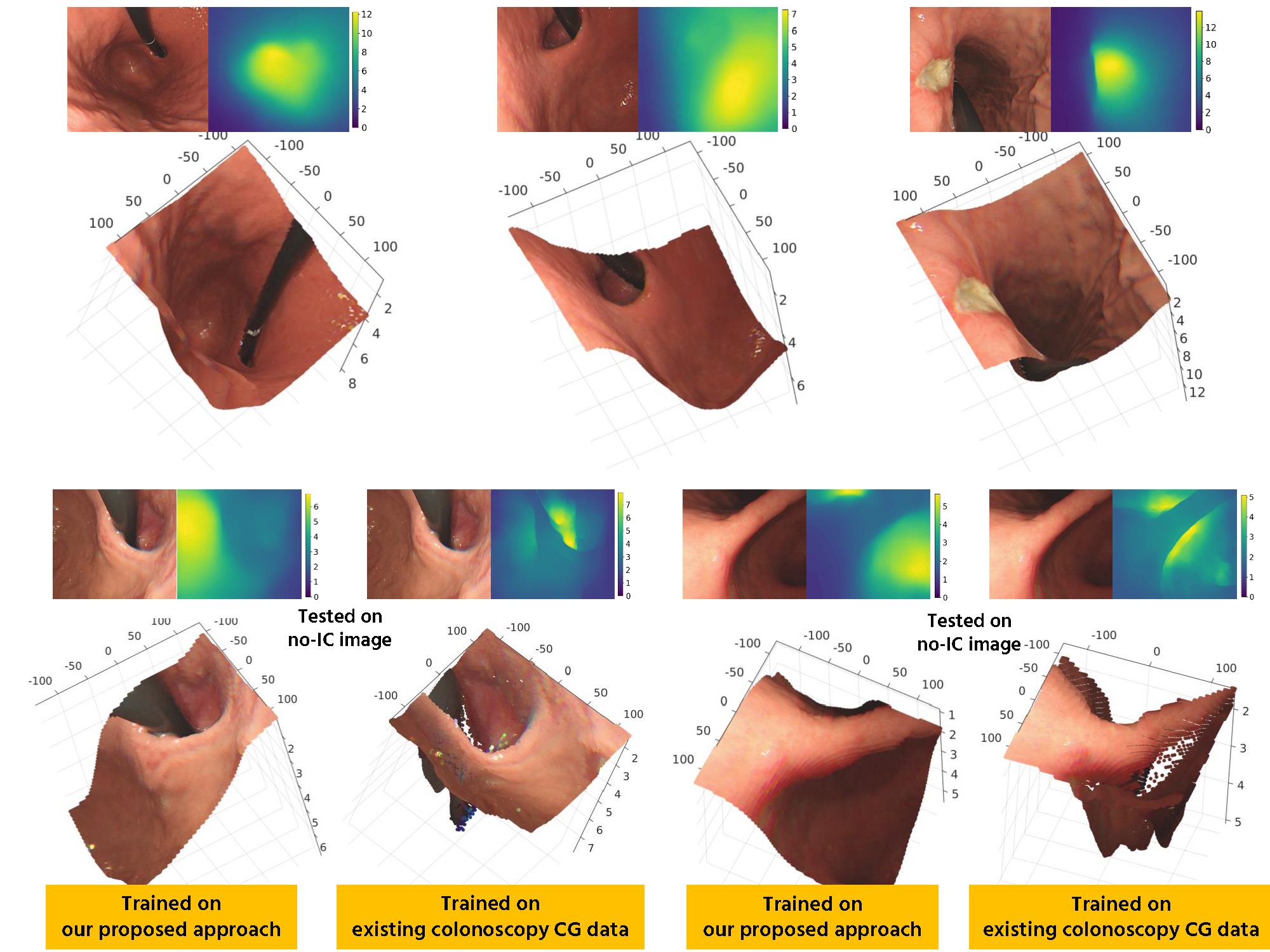
Monocular depth estimation training Even tough our main goal is to predict the depth from conventional endoscopic images without IC dye, which can be achieved by training using virtual no-IC images only, we are also aware that chromoendoscopy with IC dye is widely applied in gastroendoscopy. Because of that, we use both real IC and
virtual no-IC images and mix them into the training set to make our network applicable to both data types in the application phase.